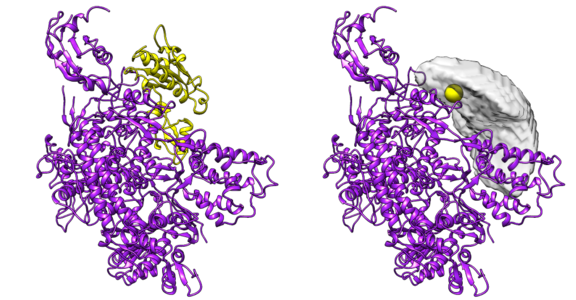
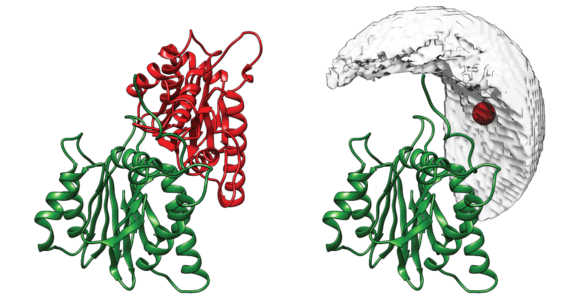
DisVis visualizes the accessible interaction space!
DisVis allows you to visualize and quantify the information content of distance
restraints between macromolecular complexes.
It performs a full and systematic 6 dimensional search of the three translational and rotational degrees of
freedom to determine the number of complexes consistent with the restraints. In addition, it outputs the
percentage of restraints being violated and a density that represents the center-of-mass position of the
scanning chain corresponding to the highest number of consistent restraints at every position in space.
In order to constantly improve our service and give you the best experience, we would really appreciate if you could take the time to complete our short online survey (~5min) available here: https://goo.gl/MYBt9R
DisVis webserver
REGISTRATION: To use the DisVis server you must have registered for an account. If you do not have an account yet you can register here
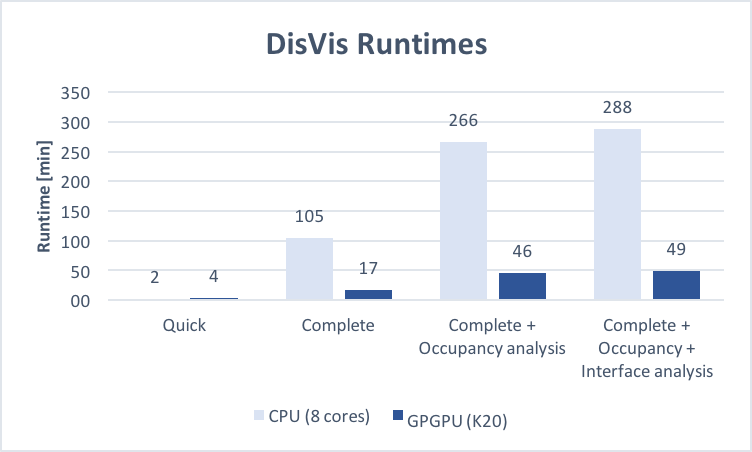
Submit your job to:Runtimes
DisVis Webinar
Reference for use of the server
- R.V. Honorato, P.I. Koukos, B. Jimenez-Garcia, A. Tsaregorodtsev, M. Verlato, A. Giachetti, A. Rosato and A.M.J.J. Bonvin (2021). Structural biology in the clouds: The WeNMR-EOSC Ecosystem. Frontiers Mol. Biosci., 8, fmolb.2021.729513.
- G.C.P. van Zundert, M. Trellet, J. Schaarschmidt, Z. Kurkcuoglu, M. David, M. Verlato, A. Rosato and A.M.J.J. Bonvin (2016)
The DisVis and PowerFit web servers: Explorative and Integrative
Modeling of Biomolecular Complexes.
J. Mol. Biol., Advanced Online Publication. - G.C.P. van Zundert and A.M.J.J. Bonvin (2015) DisVis: Quantifying and visualizing accessible interaction space of distance-restrained biomolecular complexes." Bioinformatics 31, 3222-3224.
The FP7 WeNMR (project# 261572), H2020 West-Life (project# 675858), the EOSC-hub (project# 777536) and the EGI-ACE (project# 101017567) European e-Infrastructure projects are acknowledged for the use of their web portals, which make use of the EGI infrastructure with the dedicated support of CESNET-MCC, INFN-LNL-2, NCG-INGRID-PT, TW-NCHC, IFCA-LCG2, UA-BITP, TR-FC1-ULAKBIM, CSTCLOUD-EGI, IN2P3-CPPM, SURFsara and NIKHEF, and the additional support of the national GRID Initiatives of Belgium, France, Italy, Germany, the Netherlands, Poland, Portugal, Spain, UK, Taiwan and the US Open Science Grid.
Disvis software
You can also install DisVis directly on your computer. Source code is available under Apache 2.0 license here: Github::DisVis